When: January 24, 2022
Where: FEMS Microbiology Reviews
Research Theme:
Data Life Cycle and Curation
DART Faculty Participants:
David Ussery, UAMS
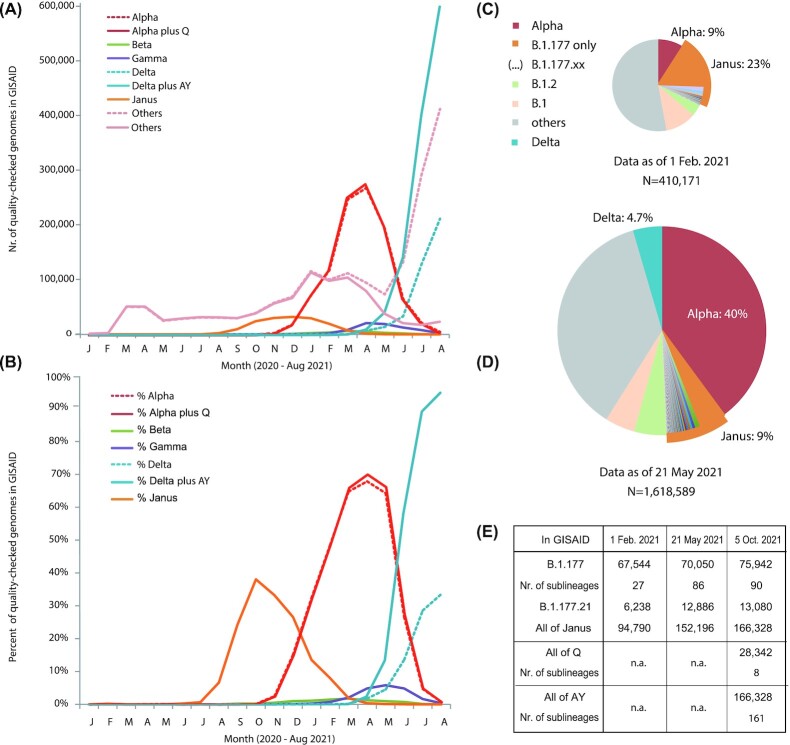
A team of researchers led by Professor David Ussery at the University of Arkansas for Medical Sciences is using high-throughput computing to help in sequencing and analysis of SARS-CoV-2 genomes within Arkansas and comparing these to millions of other Covid-19 genomes sequenced from around the world. The work performed by this team led to the discovery that the COVID-19 virus has limitations on the number of possible mutations, a significant result that will contribute to the global knowledge base needed to combat COVID-19 and similar viruses. The resulting publication is cited below.
Genomic epidemiology uses sequences of pathogens (including viruses like Covid-19) to monitor their spread and how they change with time and geographic locations. This is extremely helpful for Public Health, in terms of determining the safety and spread of a given outbreak. We have sequenced more than a thousand Arkansas Covid-19 genomes, including the first Omicron sequence in the state (this made the front page of the Arkansas Democrat Gazette on 18 December 2021), and are currently sequencing many of the ‘new Omicron variant’ BA.2 that is spreading in Arkansas. We have examined more than a million SARS-CoV-2 genome sequences, sorted by collection date, to follow the various waves of Covid-19 infections across the state (and around the world). Based on computational analysis of these millions of genomes, we have found that the mutational repertoire of SARS-CoV-2 is limited[1], and it is likely that the current vaccines will continue to be effective in preventing further outbreaks.
This work is partially supported by NSF award OIA-1946391, Data Analytics that are Robust and Trusted (DART). Dr. Ussery serves as a member of the project’s leadership team and is engaged in a number of research activities for the project.